 With RNA-based detection, the RNA is first converted into complementary DNA (cDNA) by reverse transcription. With DNA-based detection, this step is not required. Every EURORealTime test kit contains all PCR reagents ready for use, including reverse transcriptase and/or DNA polymerase and the specific primers and probes. In this way, the number of pipetting steps is reduced to a minimum: the ready-for-use PCR reagents are simply pipetted together and then the DNA/RNA is added.
With RNA-based detection, the RNA is first converted into complementary DNA (cDNA) by reverse transcription. With DNA-based detection, this step is not required. Every EURORealTime test kit contains all PCR reagents ready for use, including reverse transcriptase and/or DNA polymerase and the specific primers and probes. In this way, the number of pipetting steps is reduced to a minimum: the ready-for-use PCR reagents are simply pipetted together and then the DNA/RNA is added.
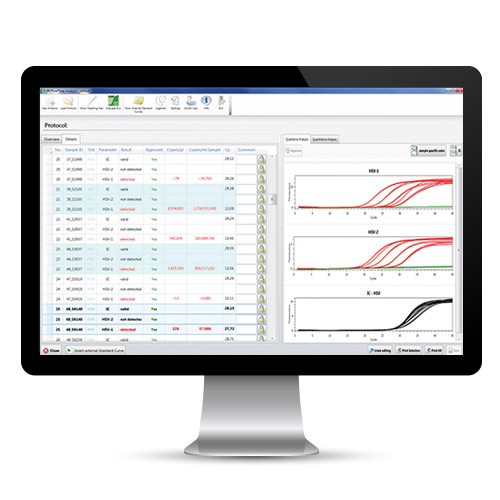
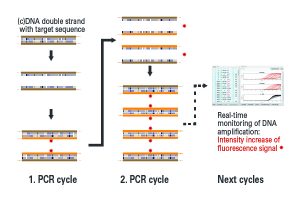
The relevant characteristic DNA/cDNA sequences are amplified millionfold by the polymerase chain reaction (PCR). Here, two starter DNA molecules (primers) define the region that is to be copied. If the sample contains the corresponding DNA sequence (target sequence), the primers can bind to it and a copy of the target sequence is produced. This reaction is repeated many times so that the DNA region between the primers is copied exponentially. During each PCR cycle, specific fluorescence-labelled DNA probes also bind to the target sequence. These only cause a fluorescence signal when the DNA has been amplified. At the end of a PCR cycle, the fluorescence intensity is measured, so that the amplification of the DNA can be followed in real time. If the target sequences are not present in the sample, the primers and probes cannot bind to them, the DNA is not amplified, and the fluorescence signal does not increase. By including the corresponding quantification standards, this method also allows the quantification of the DNA in the original sample. Moreover, different DNA sequences can be detected in one reaction by using several fluorescence dyes with different excitation and emission wavelengths.